Introduction:
This service is designed to give you the identifications and quantifications of N-glycosylated peoteins/peptides in your complex biological samples. Glycosylated proteins/peptides will be enriched using hydrazide and then subject to downstream proteome identification or quantification through Mass Spectrometry Analysis. Your samples (several test groups and a control group) will be digested with Trypsin, and peptides will be enriched using hydrazide chemical, and treated by PNGase-F, followed by chemical labeling by either iTRAQ or TMT labeling method if a quantitative analysis is desired. Enriched (with or without labeling) peptideswill be cleaned using SCX HPLC and a bRPLC fraction to further generate the 2nd dimansion of separation which has been proved to significantly increase the coverage and depth of the Mass Spectrometry Analysis.
Data analysis, including data validation, visualization and quantification, are performed with commercial softwares and Complete Omics’ unique R packages and scripts. Report will be sent to you in Excel format as well as a summary in PDF format. We will also provide you any details you need for your papers’ MATERIALS AND METHODS section. We will make sure you understand your result and help you with your paper writing with free follow-up services.
Sample types we accept:
1, 2D gel spots, SDS-PAGE bands
2, Cell Lysates and Tissue Lysates
3, Biofluids, such as plasma*, serum*, saliva, tear, etc.
*We provide High Abundance Protein Depletion Service to significantly (100-500 folds) increase the depth of your biofluid proteomics analysis by removing top abundant proteins from your samples. Read more for details.
4, FFPE slide/ FFPE extract
5, Customized sample types (please contact us to discuss)
Complete360® Enables Deep Biomarker Profiling in Groundbreaking iPSC CAR‑NK Cell Therapy Study Published in Cell
June 25, 2025 | BALTIMORE – We are thrilled to share that a major milestone in cell therapy has just been published in Cell, showcasing the world’s first-in-human clinical study of…
Complete Omics Makes Landmark Debut at ASMS 2025
June 5, 2025 | BALTIMORE -Complete Omics Inc. is proud to announce a successful debut at the American Society for Mass Spectrometry (ASMS) 2025 Annual Meeting, held in Baltimore, Maryland.
Milestone! | Complete Omics Inc. Secures California Clinical Laboratory License, Expanding Access to Cutting-Edge Clinical Proteomics Testing
Feb 20, 2025 | BALTIMORE – Complete Omics Inc., a leader in clinical proteomics and multi-omics molecular diagnostics, proudly announces its recent achievement of the California Clinical Laboratory License. This…
Some of our impacts
2023--Closing latest financing round with QiMing Venture Partners
Complete Omics proudly announces its latest financing round's success, significantly backed by Qiming Venture Partners, China's top healthcare VC. This achievement is particularly remarkable given the current economic climate, where securing investment has become increasingly challenging, especially in the healthcare and pharmaceutical sectors. Our team's dedication and resilience have been key in reaching this pivotal moment for our company. Our collaboration with Qiming, known for investing in industry leaders like Xiaomi (1810.HK), Meituan (3690.HK), and Bilibili (BILI), signals strong confidence in our mission and future. This investment propels us into new realms of innovation in clinical proteomics and molecular diagnostics, underscoring our commitment to transforming healthcare. It positions us among influential firms, promising a future of growth and breakthroughs despite the economic slowdown. We're excited for what's ahead and invite our community to join us in this journey towards significant healthcare advancements. Together, we're poised to redefine the landscape of personalized medicine. (Ref. 1, 2).A Proud Partner of QiMing VC2022--Direct Identification and Quantification of Neoantigens from Minute Amount of Clinical Biopsy Sample --- published on Cancers
In recent years, neoantigens are becoming popular cancer therapeutic targets under intensive studies by almost all major oncology pharmaceuticals. However, who are the target patients? Does the patient with a "correct" mutation and a "correct" HLA allele indeed present the "correct" neoantigen? Is this individual's neoantigen copy number high enough for immunotherapy? AI predictions based on NGS genomic information have been proven incapable of answering these questions. Immunopeptidome through mass spectrometry is dominated by disease-irrelevant peptide sequences. There is no existing way to identify and quantify neoantigens from a minute amount of clinical biopsy sample, such as 50 mg tissue or less. Valid-NEO is developed to fit this demanding clinical need through combining our proprietary multi-omics platforms including NGS-based ultra-rare mutation calling technique, DEEPER-SeqS, and our unique clinical proteomics platform, Complete360®, with additional hardware innovations (Ref. 1).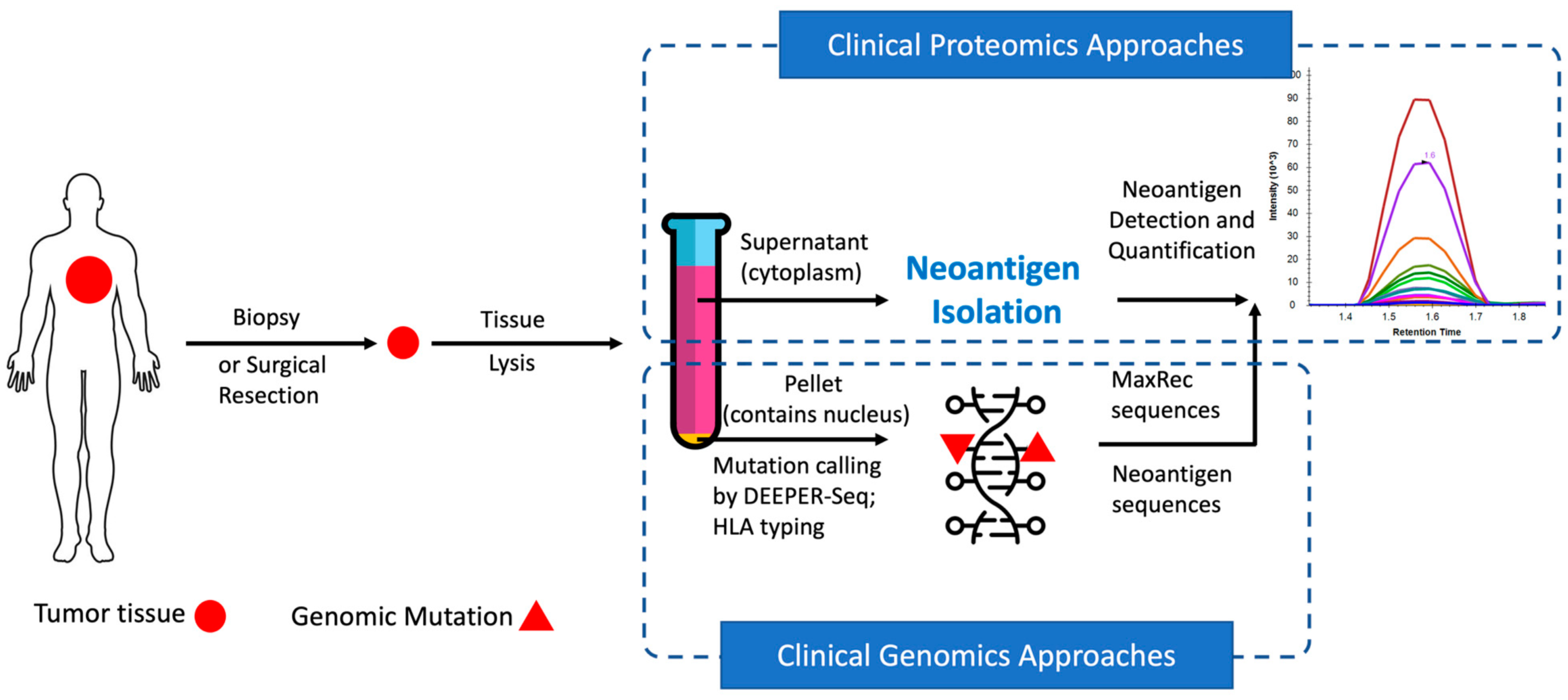 Valid-NEO: Multi-omics Pipeline for Neoantigen Assays
Valid-NEO: Multi-omics Pipeline for Neoantigen Assays2022--Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) --- published on Science Advances
Human endogenous retroviruses (HERVs) represent 8% of the human genome. Working with, ErVaccine Technologies, our scientists identified neoantigens encoded by HERVs across a broad spectrum of cancers, and this finding may help enable next-generation therapeutic vaccines and cellular immunotherapies targeting these so-called “unconventional” tumor antigens (Ref 1). These antigens are shared by different tumor types. They would prove useful as personalized cancer therapeutic targets for a large number of patients. Along the side when we are keep on improving and developing our own disease detection and companion diagnostic pipelines, we are excited to be working with a large number of collaborators to implement our multi-omics platforms in their different clinical and basic research projects.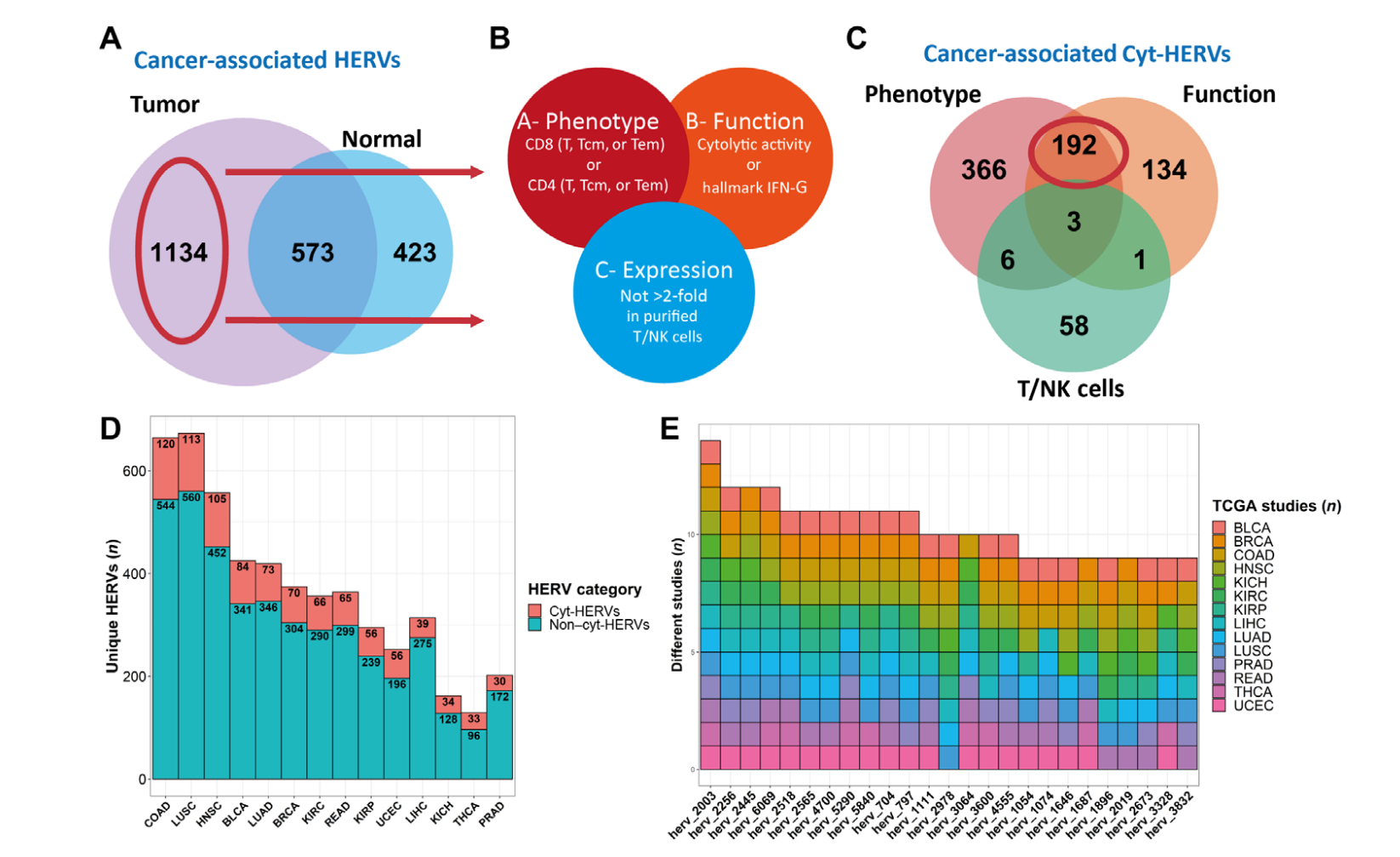 Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) — A new class of cancer therapeutic targets
Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) — A new class of cancer therapeutic targets2021--Therapeutic Neoantigens Encoded by Oncogene K-Ras --- published on Science Immunology
K-Ras is one of the most highly mutated oncogenes in cancers. The neoantigens encoded by K-Ras can be presented by many different types of cancer cells. Here we utilized our multi-omics neoantigen validation pipeline to detect and quantify K-Ras neoantigens from a variety of cancer samples (Ref 1.). We found that the K-Ras as well as several other oncogenes can be presented on cancer cell surface, but at extremely low copy numbers. We adopted a variety of internal control system to measure their abundance down to <1 copy per cell level. To come up with an actionable strategy, the therapeutic team at JHU developed bispecific antibodies that can target the neoantigens we identified, and deliver dramatic therapeutic effects to mice. These findings are significant in the way that it opens a gate to developing pan-cancer immunotherapeutic agents that can treat a large number of patients sharing cancer hotspot mutations. Identifying such neoantigens is the first step in this campaign and is being accomplished by Complete Omics Inc.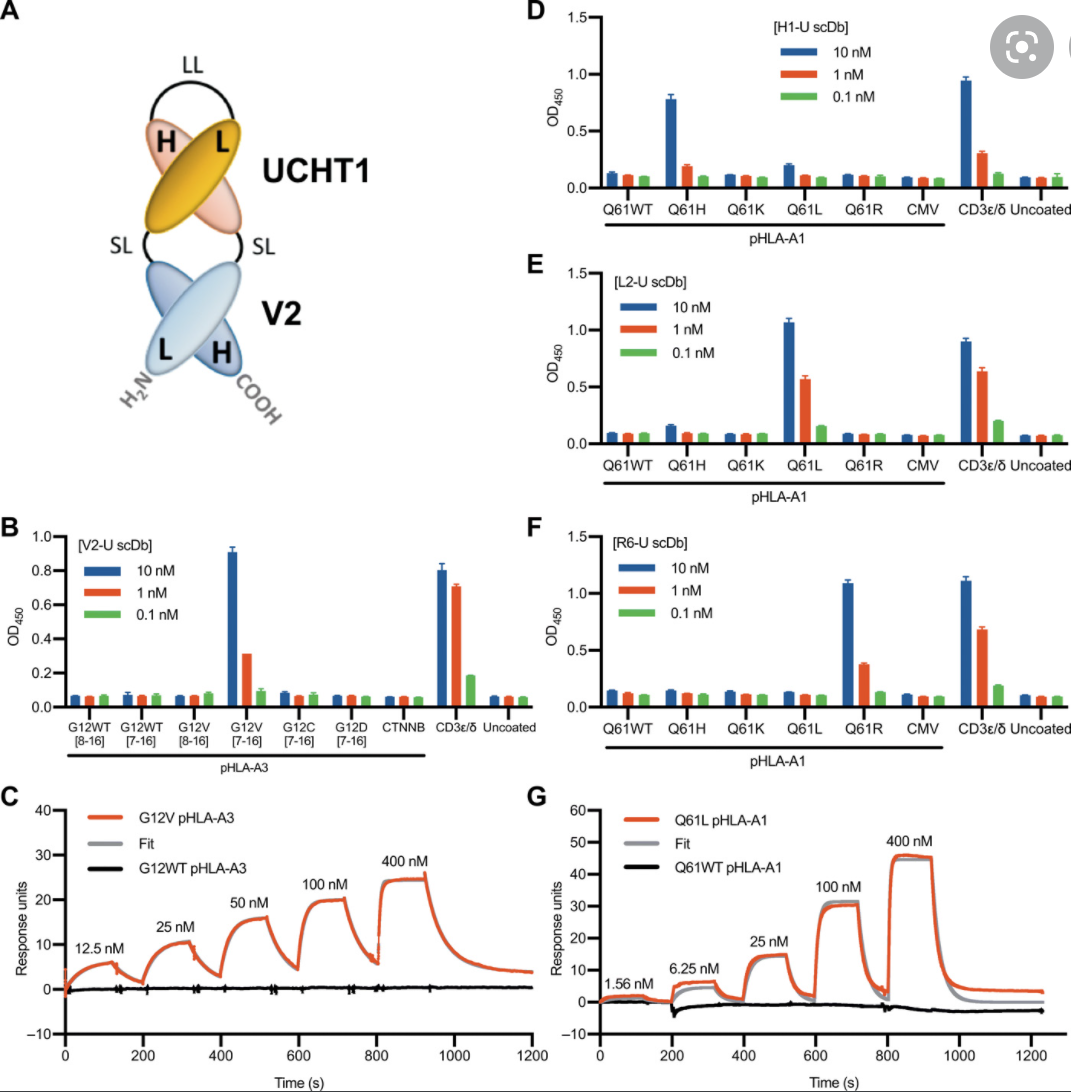 K-Ras Neoantigen Identified for Personalized Cancer Therapeutics
K-Ras Neoantigen Identified for Personalized Cancer Therapeutics2021--Direct Quantification of Neoantigens Like Never Before --- published on Science
Genetic changes in human genome are the driving force for all cancers. Different patients have different sets of mutation profiles even for the patients who all have the same disease. For decades, doctors, cancer researchers, and pharmaceutical companies have been working tirelessly trying to find a way to treat each person's unique disease in a highly personalized way that will reach the maximum treatment efficacy with the lowest side effects. Complete Omics, working with leaders in cancer therapeutics, has developed pipelines based on our multi-omics techniques through which we clearly observe and quantify personalized therapeutic targets encoded by the most frequently mutated tumor suppressor gene TP53. We validated and quantified the TP53 neoantigens on the surfaces of cancers and provided information to healthcare providers to support their decision on if or not to adopt a highly personalized cancer treatment targeting these neoantigens and when to use it (Ref 1). These findings provided the 1st-hand evidence for cancer therapeutics without the uncertainty that comes with predictions.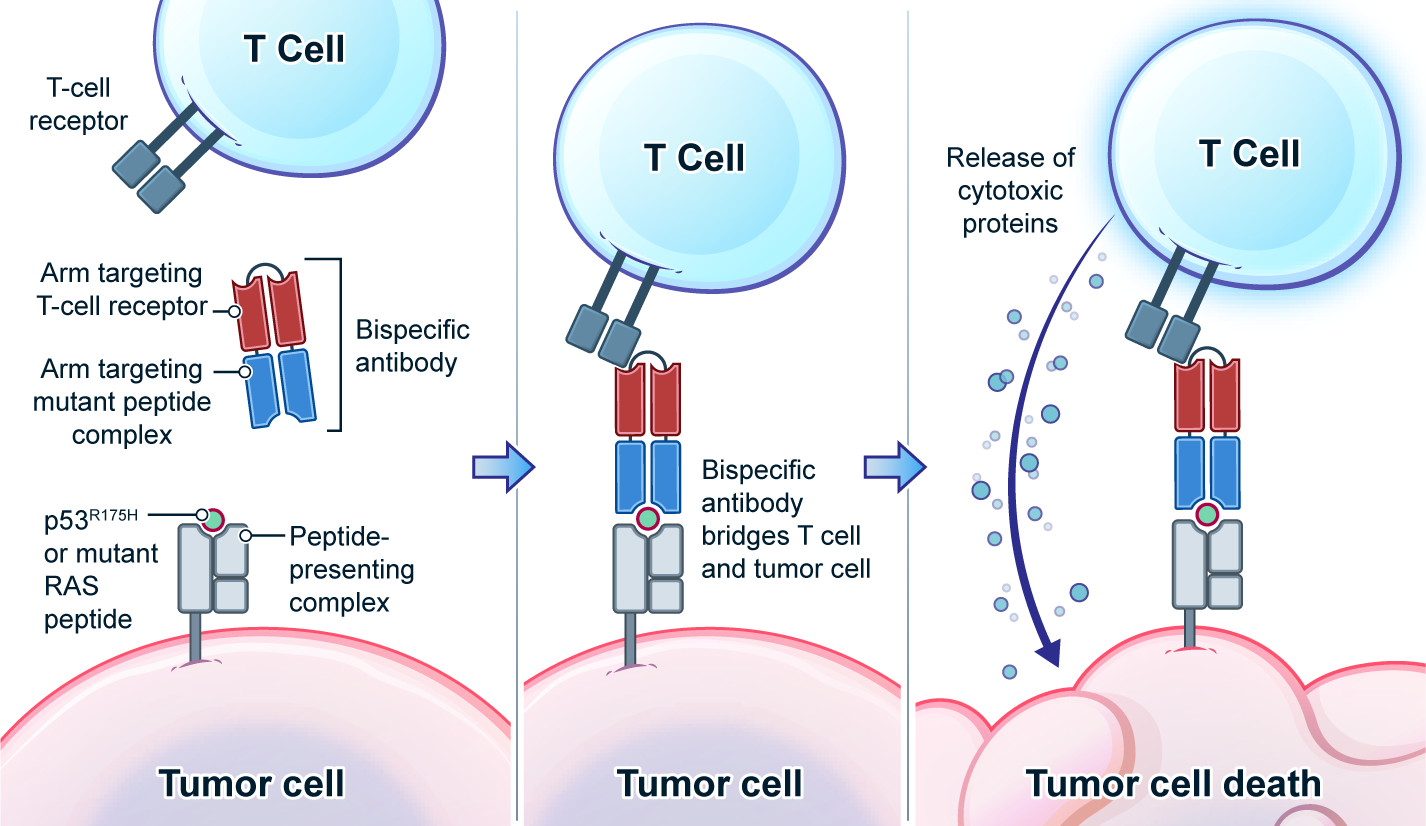 Prediction-FREE Neoantigen Validation Enables TP53-targeted Personalized Cancer Therapeutics
Prediction-FREE Neoantigen Validation Enables TP53-targeted Personalized Cancer Therapeutics