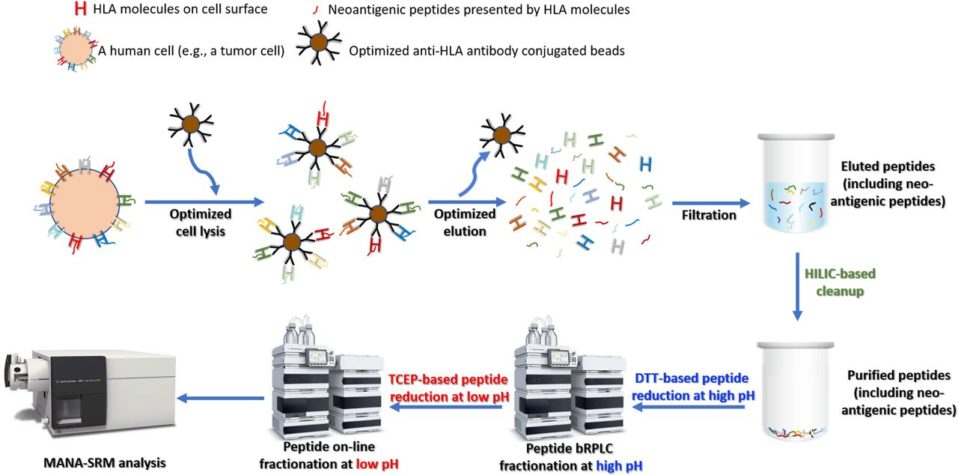
2019–First Pipeline for Direct Detection and Quantification of Neoantigens — published on Cancer Immunology Research
Cancer-linked genetic mutations can code for mutant proteins, which can then be processed by proteasomes into peptides that are presented by human leukocyte antigen (HLA) molecules, triggering the body’s immune response. The idea that such mutant peptides can trigger an immune response is fundamental to immunotherapies like checkpoint inhibitors as well as cancer vaccines that present the body with these peptides to generate an immune reaction. However, while the rise of next-generation sequencing has allowed researchers to identify a large number of cancer-linked mutations, actually detecting these mutation-associated neo-antigens, or MANAs, at the peptide level remains difficult. The fact that a mutant is present at the genetic level does not mean it will be produced at the protein level, and, even if it is, that is no guarantee that it will be processed and presented by HLA molecules. This has proved a challenge for, for instance, personalized cancer vaccine development. Our technology (Ref. 1) provide the 1st pipeline for detecting personalized cancer therapeutic targets. Our method has been reported by public media and has a significant impact in cancer research (Ref. 2).